Molecule using 3D spheres¶
Tags: sphere, arcball
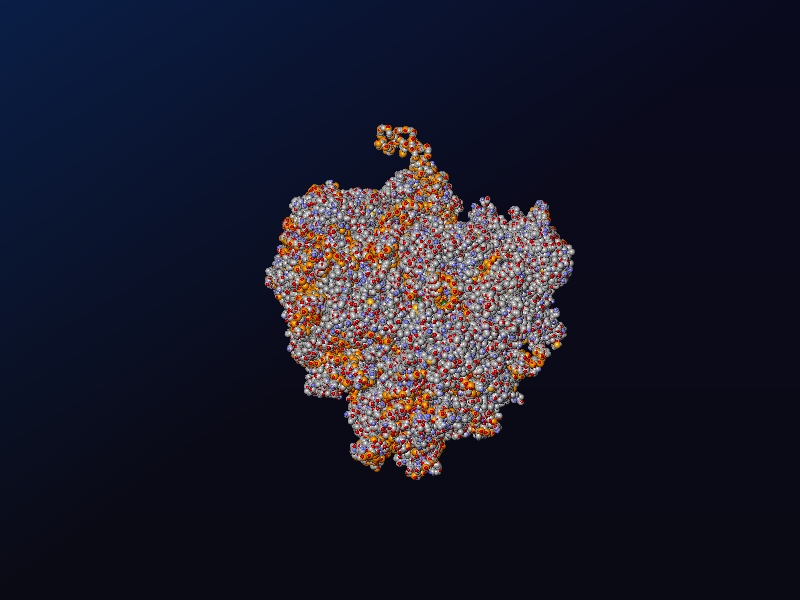
import glob
from pathlib import Path
import numpy as np
import datoviz as dvz
def load_data():
# comes from: https://www.rcsb.org/structure/6QZP
import MDAnalysis as mda
ROOT_DIR = Path(__file__).resolve().parent.parent.parent
files = sorted(glob.glob(str(ROOT_DIR / 'data/misc/molecule/6qzp-pdb-bundle*.pdb')))
universes = [mda.Universe(f) for f in files]
position = np.concatenate([u.atoms.positions for u in universes], axis=0)
element_colors_u8 = {
'H': (255, 255, 255),
'C': (200, 200, 200),
'N': (143, 143, 255),
'O': (240, 0, 0),
'S': (255, 200, 50),
'P': (255, 165, 0),
'Mg': (42, 128, 42),
'Zn': (165, 42, 42),
}
color = np.concatenate(
[
np.array([element_colors_u8[atom.element] for atom in u.atoms], dtype=np.uint8)
for u in universes
],
axis=0,
)
color = np.concatenate([color, np.full((len(color), 1), 255, dtype=np.uint8)], axis=1)
vdw_radii = {
'H': 1.20,
'C': 1.70,
'N': 1.55,
'O': 1.52,
'S': 1.80,
'P': 1.80,
'Mg': 1.73,
'Zn': 1.39,
}
atomic_radii = np.concatenate(
[np.array([vdw_radii[atom.element] for atom in u.atoms]) for u in universes]
)
size = atomic_radii.astype(np.float32)
return position, color, size
# # Save the data file
# p, c, s = load_data()
# np.savez(ROOT_DIR / 'data/misc/molecule/mol.npz', position=p, color=c, size=s)
# Load the data
data = np.load(dvz.download_data('misc/molecule/mol.npz'))
position = data['position']
color = data['color']
size = data['size']
N = len(position)
print(f'Loaded {N} atoms')
# Normalization.
position -= position.mean(axis=0)
position /= np.max(np.linalg.norm(position, axis=1))
size = 0.005 + 0.015 * (size - size.min()) / (size.max() - size.min()).astype(np.float32)
app = dvz.App()
figure = app.figure()
panel = figure.panel(background=True)
arcball = panel.arcball()
camera = panel.camera()
visual = app.sphere(
position=position,
color=color,
size=size,
lighting=True,
)
panel.add(visual)
app.run()
app.destroy()