Volume Visual¶
The volume visual displays 3D scalar fields using ray-based volume rendering. It supports NDC-aligned bounding boxes, axis permutation, basic colormapping, and limited transfer functions.
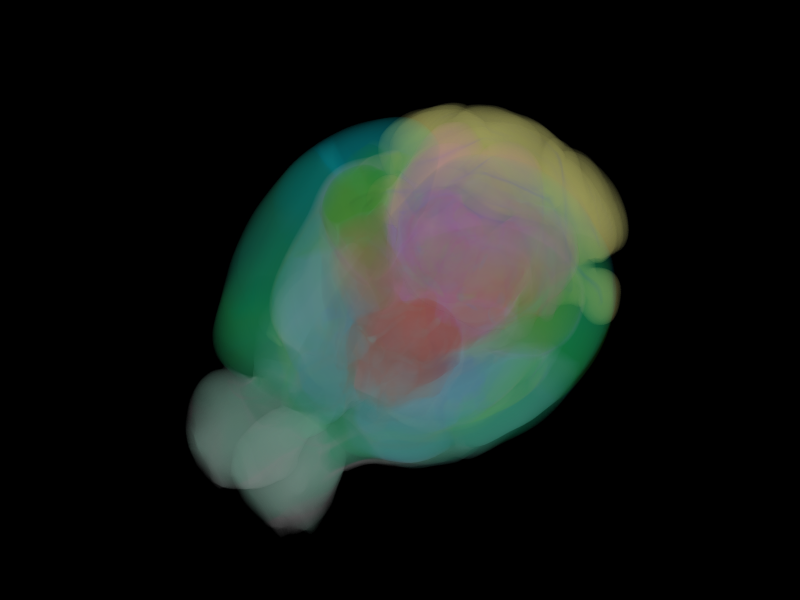
Overview¶
- Renders 3D scalar data in a box defined by NDC bounds
- Basic support for colormapping and transfer functions
- Texture-based rendering with customizable axis permutation
- Useful for initial previews of volumetric data
Warning
Volume rendering is currently a basic implementation. It is neither highly efficient nor visually polished yet. Slicing, advanced transfer functions, and performance improvements are planned for future versions.
When to use¶
Use the volume visual when:
- You want to visualize 3D data like MRI, CT, simulations
- You need a fast volumetric rendering preview
- You are working within NDC and want basic shading support
Properties¶
Options¶
| Option | Type | Description |
|---|---|---|
mode |
enum |
Color mode |
Per-item¶
| Attribute | Type | Description |
|---|---|---|
bounds |
3 × (2,) float |
Bounding box in NDC: xlim, ylim, zlim |
texcoords |
2 × (3,) float |
UVW texture coordinates at volume corners |
Per-visual (uniform)¶
| Parameter | Type | Description |
|---|---|---|
permutation |
(3,) int |
Axis order of the 3D volume texture (default (0,1,2)) |
slice |
int |
Slice index (not implemented yet) |
transfer |
vec4 |
Transfer function parameters (limited) |
texture |
texture | 3D volume data (e.g. density, intensity) |
Creation¶
To create a volume visual:
visual = app.volume(mode='colormap')
Supported modes:
| Mode | Description |
|---|---|
colormap |
Apply a colormap to a single-channel 3D texture |
rgba |
Use a 4D texture (RGBA channels for each voxel) |
Bounds and texture mapping¶
You must define the bounds of the volume in NDC:
visual.set_bounds(xlim, ylim, zlim)
Then set the texture coordinates:
visual.set_texcoords(uvw0, uvw1)
This maps the 3D texture onto the NDC bounding box.
Axis permutation¶
To reorder the axes of the 3D texture (e.g. for wvu-ordered data), set:
visual.set_permutation((2, 1, 0)) # wvu
This ensures correct alignment of the volume with your data orientation.
Transfer function¶
You can supply a transfer function parameter (currently very limited):
visual.set_transfer((0.3, 0.5, 0.8, 1.0)) # placeholder values
More expressive transfer functions (e.g. opacity maps, histograms) are planned.
Example¶
import gzip
import numpy as np
import datoviz as dvz
def load_mouse_brain():
filepath = dvz.download_data('volumes/allen_mouse_brain_rgba.npy.gz')
with gzip.open(filepath, 'rb') as f:
return np.load(f)
volume = load_mouse_brain()
shape = volume.shape
dtype = volume.dtype
D, H, W = shape[:3]
scaling = 1.0 / D
x, y, z = W * scaling, H * scaling, 1
app = dvz.App()
figure = app.figure()
panel = figure.panel()
arcball = panel.arcball(initial=(-2.25, 0.65, 1.5))
camera = panel.camera(initial=(0, 0, 3))
texture = app.texture_3D(volume, shape=(W, H, D), interpolation='linear')
visual = app.volume(
bounds=((-x, +x), (-y, +y), (-z, +z)), texture=texture, mode='rgba', transfer=(0.2, 0, 0, 0)
)
panel.add(visual)
app.run()
app.destroy()
Summary¶
The volume visual provides a starting point for 3D scalar field rendering.
- ✔️ Bounding box in NDC
- ✔️ Texture mapping with colormap
- ✔️ Axis reordering support
- ❌ No slicing or rich transfer functions (yet)
See also: